Previous Research
Auxin and the Nuclear Pore Complex

In the first few years of my time in the Estelle lab I investigated the role of the nuclear pore complex in the auxin response in Arabidopsis thaliana. We identified two genes from a suppressor screen of the auxin response mutant axr1. SAR1 and SAR3 (Suppressor of AXR1) encode the nuclear pore proteins NUP160 and NUP96 respectively.
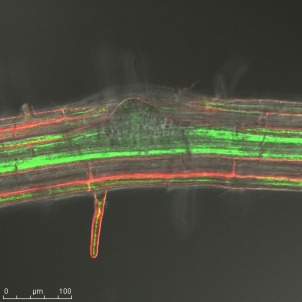
Sar1 and sar3 single mutant plants have a modest phenotype but I discovered that sar1sar3 seedlings have a severe developmental growth phenotype . In these plants both messenger RNA and an AuxIAA protein are incorrectly retained within the nucleus which would result in a wide array of developmental defects. This is the work I took on with me to when I started my new lab at Liverpool University.
Phenotype of sar1sar3 mutant plants
Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M (2006) The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE Proteins Are Nucleoporins with an Important Role in Hormone Signaling and Development. Plant Cell.
| geraintparryplantcellpdf.pdf | |
| File Size: | 751 kb |
| File Type: | |
Elucidating the role of the Arabidopsis Auxin receptor
The TIR1/AFB auxin receptor gene family contain four closely related proteins. Reducing the number of each of these proteins in Arabidopsis results in increased resistance to exogenous applied auxin. Ultimately a high proportion of triple tir1afb2afb3 and quadruple tir1afb1afb2afb3 seedlings are lethal, being unable to proceed past the early seedling stage. My research involves investigating the contribution of each family member in the seedlings response to auxin.
The role of the microRNA393
MicroRNAs are a small RNA species that can control the expression of protein coding genes. The miR393 has been shown to influence the function of the TIR1/AFB gene family in the plants response to bacterial infection (Navarro et al, 2006). My studies suggest that miR393 plays a subtle role in the control of the auxin response. However the expression domain of miR393 suggests that it is involved in the events that control lateral root development.
GFP expression driven by the miR393 promotor in a cell file surrounding an emerging lateral root.
Binding Studies
The four closely related TIR1/AFB auxin receptors bind to AuxIAA proteins which precedes the ubiquitination and subsequent degradation of these proteins. There are 29 different AuxIAA proteins in Arabidopsis and we used a variety of methods including the Yeast Two Hybrid system to investigate the interactions between auxin receptors and AuxIAA proteins. This strategy has enabled us to draw conclusions about the functional significance of specific interactions between these protein families.
Geraint Parry, L.I. Calderon-Villalobos, M. Prigge, B. Peret S. Dharmasiri, H. Ito, E. Lechner, W.M. Gray, M. Bennett, and M. Estelle (2009): Complex Regulation of the TIR1/AFB family of Auxin Receptors. PNAS 106: 22540
| geraintparrypnas.pdf | |
| File Size: | 1196 kb |
| File Type: | |
Pharmalogical control of auxin transport
The hormone auxin is transported through plant cells through the activity of influx and efflux carrier proteins. During my PhD I studied the role the novel chemicals 1-NOA, CHPAA and Naproxen play in the inhibition of these carrier proteins.
My research showed that 1-NOA and CHPAA could phenocopy the influx carrier protein mutant aux1 whilst Naproxen acts as a highly specific auxin efflux inhibitor. The identification of the role of 1-NOA compound has allowed subsequent research into the role auxin transport in meristem development.
Parry G, Delbarre A, Marchant A, Swarup R, Napier R, Perrot-Rechenmann, Bennett M (2001) Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1 . The Plant Journal
| geraintparryplantjournal.pdf | |
| File Size: | 220 kb |
| File Type: | |
Histone Modification and DNA replication
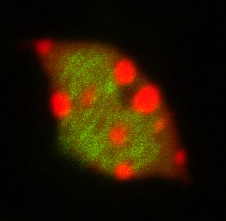
The picture to the left shows chromocenter (red) and H3K27me1 (green) staining in the leaves of ATXR5 OX nuclei.
The precise mechanisms by which ATXR5 and ATXR6 act remains elusive. My project investigated the mechanisms by which these proteins function and involved studying histone modifications, expression patterns and involvement of small RNAs in the regulation of many genetic elements. This used both conventional molecular biology techniques as well as whole genome analyses. During this time I learnt a range of new skills that were of great benefit as I started to develop my own independent research.
